Bacterial Growth & plasmid
preparation *
STORAGE OF E. COLI STRAINS *
"Slopes" *
"Stabs" *
Glycerol Cultures *
HMFM Cultures *
Glycerol L-Broth Plates *
GENERAL BACTERIAL GROWTH *
L-Broth *
L-Broth Plates *
2x YT media: *
1x YT media: *
YT plates: *
YT Soft agar: *
Glucose minimal plates: *
Cultures *
STOCK SOLUTIONS FOR PLASMID PREPARATIONS *
Antibiotics: *
For cleared lysate preparations: *
For NaOH/SDS preparations: *
LARGE SCALE PLASMID CULTURE-CLEARED LYSATE PREPARATION
*
SMALL SCALE PLASMID CULTURE-CLEARED LYSATE PREPARATION
*
SMALL SCALE PLASMID (COSMID) PREPARATION *
LARGE SCALE PLASMID (COSMID) PREPARATION *
ISOLATION OF MINI-PREP PLASMID DNA FOR SEQUENCING *
Primer annealing: *
Alternative rapid denaturation & annealing: *
FURTHER PLASMID PURIFICATION *
A. On Sucrose Gradients *
B. On Caesium Chloride-Ethidium Bromide Gradients *
___________________________________________________________________________
Bacterial Growth & plasmid preparation
STORAGE OF E. COLI STRAINS
Note. For plasmids 1 to 10 ug of DNA should be stored at -20oC or -80oC in TE for
ultimate safety.
Strains can be stored as colonies on normal L-broth plates
for about 2 months at 4oC.
"Slopes"
For longer storage at 4oC and as a practical
source of strain, bacteria are grown up as a lawn on "slopes",
~10 ml tubes in which 2 ml (antibiotic) L-broth agar is allowed
to set while tube laying nearly flat. Produces a large surface
culture.
"Stabs"
For longer storage and dispatch "stabs" are used.
Small glass vials half filled with 0.5 to 1 ml (antibiotic) L-broth
agar. Bacteria are stabbed deep into the agar and allowed to grow
under ideal conditions 0/N, then stored at 4oC.
Viable for up to 1 year.
Glycerol Cultures
To 0.5 to 1 ml of a log phase or saturated bacterial culture
add ~1.2 volumes of sterile 80% glycerol in H20 (autoclaved).
Store at -20oC. If stored at 70oC
culture will freeze and should be thawed only once. Viable for
about a year, possibly longer if freezing and thawing does not
take place.
HMFM Cultures
Hogness modified freezing medium is used for storage of cultures
at -70oC. Cultures are less affected by
freezing and thawing in this medium (10 cycles typically gives
a ten fold loss) and are viable for many years. The cultures can
be sent at R/T through mail and even refrozen on arrival.
A log phase culture (OD 660 nm (1 cm) = 0.8) or even a saturated
culture is centrifuged to harvest cells. After pouring off the
medium, cells are resuspended in sterile:
3.6 mM K2HPO4
1.3 mM KH2PO4
2.0 mM Na3citrate
1.0 mM MgSO4.7H2O
4.4% (v/v) glycerol
and kept at -70oC. NOTE do not
store at -20oC even for a short period.
Glycerol L-Broth Plates
For storage of many bacterial colonies, eg a gene bank, the
colonies plated on itrocellulose are transferred to an (antibiotic)
L-broth agar plate containing 20-30% glycerol (added before autoclaving
agar). Plates are then sealed and stored at -20oC
or -70oC. If a colony is required, filter
ONLY is transferred to (antibiotic) L-broth agar plate at R/T
and glycerol plate immediately replaced at -20oC.
After work with filter is complete it is returned rapidly to glycerol
plate in freezer.
GENERAL BACTERIAL GROWTH
L-Broth
| Tryptone (Difco) |
10 g |
|
50 g |
|
Yeast Extract
(BBL or Difco)
|
5 g |
|
25 g |
| NaCl |
5 g |
|
25 g |
| |
----- |
|
----- |
| total volume |
1 litre |
|
5 litres |
Dissolve ingredients in H2O and adjust
pH to 7.2 with NaOH (about 2 ml 5 M NaOH/litre, but depends on
tryptone and yeast extract lots). Autoclave same day.
L-Broth Plates
12.5 gm of agar (BBL or Difco) added to 1 litre fresh L-broth
and autoclaved. The agar can then be stored (mix well after autoclaving)
or used immediately. Agar broth, either remelted at 100oC
in microwave or freshly autoclaved, is cooled to ~65oC,
antibiotic added if required and poured rapidly to a depth of
~1 cm in petri dishes. 1 litre will give about 30 x 9 cm plates.
2x YT media:
| Bacto®
Tryptone |
16 g |
| Bacto®
yeast extract |
10 g |
| NaCl |
5 g |
| |
----- |
| Final volume |
1 l |
Dissolve ingredients in H2O and adjust
pH to 7.2 with NaOH.
Autoclave the same day.
1x
YT media:
| Bacto®
Tryptone |
8 g |
40 |
| Bacto®
yeast extract |
5 g |
25 |
| NaCl |
5 g |
25 |
| |
----- |
----- |
| Final volume |
1 l |
5 l |
YT
plates:
Add 15 g/litre (3 g/200 ml) Difco agar to YT media. Autoclave
the same day.
YT
Soft agar:
Add 6 g/litre to YT media (1.2 g/200 ml). Autoclave the same
day.
Glucose
minimal plates:
Autoclave (unless otherwise indicated) the following reagents
separately and cool before mixing aseptically.
| 15 g Difco Bacto®
agar in 888ml H2O |
888 ml |
| 10x M9 salts |
100 ml |
| 1 M MgSO4 |
1 ml |
| 1 M CaCl2 |
0.1 ml |
| 1% thiamine HCl, sterilised by filtration |
1 ml |
| 20% glucose |
10 ml |
| |
------- |
| Final volume |
1 l |
10x M9 Salts:
| Na2HPO4 |
|
| KH2PO4 |
|
| NH4Cl |
|
| NaCl |
|
| |
|
| Final volume |
|
Autoclave and store at 4oC.
Cultures
Large cultures are grown in smooth walled flasks containing
up to 20 to 25% total flask-volume of L-broth. Normally flasks
are incubated at 37oC and shaken at about
220 to 250 rpm.
STOCK SOLUTIONS FOR PLASMID PREPARATIONS
Antibiotics:
| Filter sterilised |
Kanamycin |
100 mg/ml H2O
(1000x stock) |
| or prepared |
Ampicillin |
50 mg/ml in H2O
(1000 x stock) |
| in sterile H2O |
Tetracyclin |
12.5 mg/ml in H2O
(1000x stock) |
| Prepared in EtOH |
Chloramphenicol |
170 mg/ml in EtOH (1000x stock) |
For cleared lysate preparations:
50 mM Tris HCl pH 8.0
500 mM EDTA ~pH 8.3
10% Triton X-100 in H2O
3 M NaAc ~pH 7.0
Isopropanol
EtOH at -20oC
80% EtOH in H2O at -20oC
1x TE (20 mM Tris HC1, 1 mM EDTA, pH 8.3, from 100 x stock)
Phenol, 0.1% 8-hydroxyquinoline, saturated with 0.3 M TNE
(stored at 4oC)
Chloroform - isoamylalcohol (24:1 v/v) (stored at 4oC)
RNAase A, 20 mg/ml in 150 mM NaAc pH 5 (made up from Sigma
best grade, boiled 5 mins and then frozen in 0.5 ml aliquots
-20oC)
0.3 M NaAc (from 3 M stock)
0.3 M NaAc/isopropanol (1:0.6)
For NaOH/SDS preparations:
50 mM glucose, 10 mM EDTA, 25 mM Tris HCl pH 8.0
0.2 M NaOH, 1% SDS
3 M KHAc pH 4.8 (KHAc dissolved initially at 6M, then titrated
to pH 4.8 with glacial acetic and finally volume adjusted to
give 3M K)
RNAase A 20 mg/ml (as above)
10x TE (from 100 x stock)
Phenol (as above)
EtOH at -20oC
80% EtOH in H2O at -20oC
0.3 M NaAc (from 3 M stock)
0.3 M NaAc/isopropanol (1:0.6)
LARGE SCALE PLASMID CULTURE-CLEARED
LYSATE PREPARATION
| pCRI |
|
Kanamycin |
100 ug/ml |
| pBR322 etc |
either |
Tetracycline |
12.5 ug/ml |
| |
or |
Ampicillin |
50 ug/ml |
2. Leave to grow overnight at 37oC.
3. Transfer an isolated colony to 10 ml antibiotic L-broth
and leave to grow O/N until saturated, at 37oC
in the shaking incubator. Use sterilin universals.
4a. Inoculate 1 litre of antibiotic L-broth with 5 to 10 ml
of the saturated culture. Leave to grow at 37oC
in the shaking incubator until the OD 600 nm (1 cm) = 1.0-1.5.
Add chloramphenicol to a final concentration of 170 ug/ml
(stock solution is 170 mg/ml in ethanol - N.B. DO NOT FLAME).
Leave in shaking incubator at 37oC for
more than 12 hours.
or
4b. Inoculate as in 4a. and grow until well after saturation,
i.e. more than 24 hours.
5. Spin in 1 litre buckets on the Sorvall RC-3B (soft stop
to avoid resuspension of bacterial pellet) for 15-20 minutes
at 4,000 rpm and 4oC.
6. Carefully pour off the supernatant not disturbing the pellet.
Stand the buckets upside down on absorbent paper to drain off
the last of the supernatant.
7. Take up the pellet in 10 ml of 50 mM Tris HCl pH 8 and
transfer to 50 ml Oakridge centrifuge tubes.
8. Add 1 ml of lysozyme solution (10 mg/ml) - freshly made
up in 50 mM Tris pH 8. Mix well - whirlimix - leave for 10 minutes
on ice.
9. Add 1 ml 0.5 M EDTA (final concentration 50 mM EDTA), mix
well but gently -roll, DO NOT SHAKE. DO NOT WHIRLIMIX. Leave
for 10 minutes on ice.
10. Add 50 ul RNase A (20 mg/ml).
11. Add 100 ul 10% Triton and mix gently, but thoroughly,
by rolling tube. Leave on ice for 20 minutes.
12. Spin on the Sorvall RC-5B, SS34 rotor for 1 to 2 hours
at 18,000 rpm and 4oC (the longer the
spin the better).
13. Pour off the supernatant quickly but smoothly into a clean
50 ml centrifuge tube. The supernatant should be clear and not
viscous. It must be removed as soon as the centrifuge stops.
The viscous pellet will start to resuspend if left for any time
at all.
14. To the cleared lysate add 1 ml 3 M NaAc and 10 ml phenol.
Seal tube with plastic cap and shake well. Spin for 15 minutes
at 10,000 rpm on the Sorvall RC-5B, HB4 rotor at R/T.
15. Remove the upper phase into a clean tube and add another
10 ml phenol to it. Mix and spin again. Repeat until no interphase
is seen.
16. Repeat with chloroform:iso-amyl alcohol (24:1) instead
of phenol.
17. Remove the upper phase into another clean 50 ml centrifuge
tube and precipitate DNA and RNA with 25 ml cold ethanol. Leave
at -20oC for more than 30 min..
18. Spin for 15 minutes at 10,000 rpm, 4oC
in Sorvall RC-5B, HB4 rotor. Pour off supernatant and lightly
vacuum dry pellet.
19. Redissolve pellet in 10 ml of 0.3 M NaAc at R/T and add
5.8 ml R/T isopropanol dropwise on whirlimixer. Leave 10 min
at R/T, then spin 15 min at 10,000 rpm and at R/T in Sorvall
RC-5B.
20. Wash once with 10 ml 0.3 M NaAc/isopropanol (1:0.6 v/v).
Spin 5 mins 10,000 rpm at R/T.
21. Wash once with 10 ml 80% ethanol (-20oC)
and spin as above.
22. Vacuum dry pellet and redissolve in 250 to 500 ul of 1x
TE.
Between 250 and 500 ug of plasmid can be expected from a litre
culture, but sometimes up to 1 mg may be obtained.
SMALL SCALE PLASMID CULTURE-CLEARED
LYSATE PREPARATION
| pCRI |
|
Kanamycin |
100 ug/ml |
| pBR322 etc |
either |
Tetracycline |
12.5 ug/ml |
| |
or |
Ampicillin |
50 ug/ml |
2. Leave to grow overnight at 37oC.
3. Transfer an isolated colony to 2 ml antibiotic-L-broth.
Leave to grow for at least 6 hours, preferably until saturated,
at 37oC in shaking incubator. Use sterilin
universals.
4. If necessary use 1 ml of this culture to make another glycerol
or HMFM culture:
Add rather more than an equal volume of autoclaved 80% aqueous
glycerol (i.e. to a final concentration of 50-60%) to 1 ml of
the culture. Store at -20oC.
5a. Inoculate up to 100 ml of antibiotic-L-broth with the
rest of the culture (1 ml) and grow at 37oC
until an OD 600 nm (1 cm) = 1.0-1.5 is reached. Add chloramphenicol
to a final concentration of 170 ug/ml and shake well for at least
a further 12 hours at 37oC. (N.B. The
chloramphenicol stock solution is made up in ethanol - DO NOT
FLAME).
or
5b. Inoculate as in 5a. and grow culture to saturation with
good aeration (more than 24 hours).
6. Harvest cells in 50 ml Oakridge tubes by spinning for 20
minutes at 4,000 rpm on Sorvall RC-3B. Carefully pour off supernatant
and leave tubes upturned on paper to drain for a few minutes.
7. Take up pellet in 0.5 ml of 50 mM Tris HC1 pH 8.
8. Transfer to Eppendorf tube.
9. Add 50 ul of lysozyme - 10 mg/ml, freshly made up in 50
mM tris HC1 pH 8 (final concentration 1 mg/ml). Whirlimix for
a couple of seconds and leave for 5 minutes on ice.
10. Add 50 ul of 0.5 M EDTA and mix well. Leave for 5 minutes
on ice.
11. Add 5 ul of RNAase A (20 mg/ml)-final concentration 100
ug/ml.
12. Add 10 ul 10% Triton and mix by gently rolling tube. Leave
on ice for 15 to 20 minutes. Should be very viscous at this stage.
13. Spin in the Sorvall RC-5B, SS34 rotor for 1 to 2 hours
at 18,000 rpm and 4oC.
14. Pour off the supernatant quickly but smoothly into a clean
eppendorf. The supernatant should be clear and NOT viscous. Any
delay in removing the supernatant will allow the viscous pellet
to resuspend.
15. Add 50 ul of 3 M NaAc and an equal volume of phenol to
the supernatant. Mix thoroughly by shaking or whirlimixing.
16. Spin in eppendorf centrifuge for 5 to 10 minutes and remove
upper phase into fresh tube.
17. Repeat extraction with phenol once or twice until no interphase
forms, then extract once with chloroform:isoamyl alcohol (24:1
v/v).
18. At this stage a small sample can be removed for a gel.
19. Precipitate plasmid and RNA with 2 vols cold ethanol and
leave at -20oC for about 30 minutes.
20. Spin in Sorvall RC-5B, HB4 rotor at 10,000 rpm for 15
min, 4oC. Remove supernatant and vacuum
dry pellet for 5 minutes.
21. Dissolve at room temperature in 0.5 ml, 0.3 M NaAc pH
7-8 and then differentially precipitate plasmid by adding 0.56
to 0.58 volumes (0.28 to 0.29 ml) of R/T isopropanol dropwise
on whirlimixer. Leave for 10 minutes at room temperature.
22. Spin in eppendorf centrifuge for 10 minutes at R/T.
23. Wash once with 0.5 ml 0.3 M NaAc/isopropanol (1:0.6 v/v),
spin 5 min R/T in eppendorf centrifuge.
24. Wash once with ice cold 80% ethanol, same spin.
25. Vacuum dry and redissolve in about 50 ul of 1x TE. Yield:
can expect ~20 ug from a 50 ml culture.
SMALL SCALE PLASMID (COSMID) PREPARATION
(Essentially as Birnboim and Doly (1979) NAR 7, 1513-1523)
1. Grow up to a 2 to 100 ml chloramphenicol amplified culture
or 2 to 25 ml saturated culture as for cleared lysate preparation.
10 ml saturated cultures can be conveniently grown in Sterilin
universal tubes and yield 20ug or more of plasmid DNA.
2. 2 ml cultures are harvested directly in eppendorf tubes
by repeated microcentrifugation. 10 ml cultures are harvested
directly in culture tubes at 3,000 rpm and 4oC
for 10 minutes in the Sorvall RC-3B using blue adaptors (conical
bottomed). Larger cultures may be harvested in 50 ml or 250 ml
tubes spinning for 15 minutes at 4000 rpm.
3. Resuspend cells in 200 ul of:
50 mM glucose
10 mM EDTA
25 mM Tris HC1 pH 8.0
Or simply 200 ul of TE and transfer to eppendorf tube on ice.
4. Add 400 ul ice cold 0.2 M NaOH, 1% SDS, mix and leave on
ice for 5 minutes exactly.
5. Add 200 ul ice cold 3 M KHAc pH 4.8 (3 M KHAc dissolved
initially at 6 M and then titrated with glacial acetic to pH
4.8, finally volume adjusted to give 3 M K).
5 ul of RNAase A (20 mg/ml) may be added at this step, but
is more effective at step 8 below. Mix and leave on ice for 15
minutes.
6. Spin in eppendorf centrifuge in cold room for 5 minutes
and remove supernatant into a fresh tube.
7. Precipitate nucleic acid by the addition of 500 ul of isopropanol
(~0.6 volumes) and centrifuge immediately in eppendorf centrifuge
for 5 minutes.
8. Remove supernatant and dissolve pellet in 300 ul 100 mM
Tris HC1, 10 mM EDTA pH 8.3 (10x TE). 5 ul of RNAase A (20 mg/ml)
may be added at this step and the solution incubated at 37oC for 10 to 30 min.
9. Add 300 ul phenol (0.1% hydroxyquinoline) saturated with
0.3 M TNE, whirlimix and spin in eppendorf centrifuge for 5 minutes.
10. Remove upper phase into a fresh tube. Depending on intended
use of prep. step (9) can be repeated once or twice, until no
interphase is seen.
11. Precipitate nucleic acid with 600 ul ice cold EtOH, mix
well and leave at 20oC for more than 10
minutes.
12. Spin in eppendorf centrifuge in cold room for 10 minutes,
pour off supernatant and allow tube to drain inverted.
At this stage the DNA is useable for restriction and nick
translation.
If it is an advantage to remove the excess RNA the following
steps may be used:-
13. Redissolve pellet in 500 ul 0.3 M NaAc ~pH 7 at R/T. Add
0.28 to 0.29 ml R/T isopropanol dropwise on whirlimix. Leave
10 minutes at R/T then spin for 10 minutes in eppendorf centrifuge
at R/T.
14. Wash with 1 ml of R/T 0.3 M NaAc/isopropanol (1:0.6 v/v)
and spin for 5 minutes as in (13).
15. Discard supernatant, add 500 ul chilled EtOH and spin
again as in (14).
16. Discard supernatant and allow tube to drain.
Plasmid DNA should be restrictable by most enzymes and may
be nick translated. 5’ labelling may also be carried out
but since a small amount of low molecular weight RNA is present
this will take up most of the label.
LARGE SCALE PLASMID (COSMID) PREPARATION
(Essentially as Birnboim and Doly (1979) NAR 7, 1513-1523)
1. Grow 500 or 1 litre culture as for cleared lysate preparation,
i.e. either using chloramphenicol amplification or growth to
saturation.
2. Harvest cells by spinning for 15-20 minutes at 4 krpm and
4oC in Sorvall RC-3B, (soft stop to avoid
resuspension of bacterial pellet).
3. Resuspend cells in 30 ml of:
50 mM glucose
10 mM EDTA
25 mM Tris HC1 pH 8.0
and transfer to 250 ml centrifuge bottles and cool on ice.
4. Add 60 ml of ice-cold 0.2 M NaOH, 1% SDS, mix well by rolling
tubes and leave on ice for 5 mins exactly.
5. Add 30 ml ice-cold 3 M KHAc pH 4.8. Mix well by gently
rolling tubes and leave for 15 minutes on ice. Finally shake
vigorously.
6. Spin at 5,000 rpm (in Sorvall RC-3B) or 10,000 rpm (in
Sorvall RC-5B, GSA rotor) for 15 mins at 4oC.
Then filter supernatant through two layers of cheese cloth (sterile
gauze) into fresh 250 ml bottles.
If the supernatant is not clear then mix solution well and
repeat the centrifugation.
7. Precipitate nucleic acid by the addition of 75 ml of isopropanol
(~0.6 vols) and centrifuge immediately at 5,000 rpm for 20 min
in Sorvall RC-3B or 10,000 rpm, 10 min in Sorvall RC-5B, GSA
rotor, setting temperature to 22oC.
8. Pour off supernatant and discard. Dissolve pellet in 10
ml of 100 mM Tris HC1, 10 mM EDTA pH 8.3 (10x TE) and transfer
to 50 ml Oakridge tube or 30ml disposable tube.
Add 50 ul of RNAase A (20 mg/ml) and leave 30 min at 37oC.
Add 1 ml 3 M NaCl (or NaAc).
9. Add 10 ml of phenol (0.1% hydroxyquinoline) saturated with
0.3 M TNE, cap tube, shake well and centrifuge for 15 min at
5000 rpm in Sorvall RC-3B (blue adaptors) or 10,000 rpm, 5-10
mins in Sorvall RC-5B, HB4 rotor.
10. Remove upper phase into fresh tube and if necessary repeat
(9) once or twice, until no interphase is seen after centrifugation.
11. Precipitate nucleic acid with 20 ml cold EtOH, mix well
and leave at -20oC for 30 mins.
12. Spin tube at 10,000 rpm for 15 min at 4oC
in Sorvall RC-5B, HB4 rotor. Pour off supernatant and discard,
leaving tube to drain for a minute.
At this stage the DNA may be redissolved for separation on
a EtBr-CsCl gradient.
If it is advantageous to remove excess RNA, something that
a CsCl gradient does only poorly, the following steps may be
used.
13. To remove bulk of RNA by differential precipitation, redissolve
pellet in 10 ml 0.3 M NaAc ~pH 7 at R/T. Add 5.6 to 5.8 ml R/T
isopropanol dropwise on whirlimix. Leave 10 min at R/T then centrifuge
for 15 mins at 10,000 rpm, R/T in Sorvall RC-5B, HB4 rotor. (Retain
S/N in case plasmid not precipitated.)
14. Wash with 10 ml of R/T 0.3 M NaAc/isopropanol (1:0.6 v/v),
spin for 5 min as in (13).
15. Discard supernatant, add 10 ml chilled EtOH and centrifuge
again as in (14) but setting temperature to 4oC.
16. Discard supernatant and leave tube to drain for a few
minutes. The plasmid may now be dissolved for immediate use or
for further purification. About 500 ug to several mg of plasmid
(100 ug for cosmids) can be expected from a 500 ml to 1 litre
culture.
ISOLATION OF MINI-PREP PLASMID DNA
FOR SEQUENCING
1. Decant 2,5 ml O/N bacterial culture into a eppendorf tube,
spin for 2 minutes. Completely remove the supernatant by aspiration.
2. Resuspend the pellet in 200 ul GTE (50mM glucose, 10mM
EDTA, 25mM Tris-HCl pH8.0) solution and vortex.
3. Add 400 ul 0.2 N NaOH/l% SDS and invert several time before
placing on ice for 5 minutes. Do not vortex!
4. Add 300 ul 3 M K acetate (pH 4,8), invert several times
and place on ice for 7 minutes. Do not vortex!
5. Centrifuge for 5 minutes and decant (simply pour) the supernatant
into another tube. (Here a phenol/chloroform followed by a chloroform
extraction can be included, but is not necessary for sequencing
reaction). Add 700 ul absolute ethanol, mix (vortex very well)
and spin immediately for 5 minutes.
6. Discard the supernatant and wash the pellet with l ml 70%
ethanol. Spin 5 minutes, drain tube well and vacuum dry the pellet.
7. Resuspend the DNA pellet in 30 ul TE buffer. For restriction
enzyme digestion take 2-4 ul (depending how big the insert is).
Primer annealing:
Take 5-8 ul of the plasmid prep., add 8-11 ul water and 4 ul
l N NaOH/0,1 mM EDTA, mix and incubate for 5 minutes at 80-99
oC, we usually use 99oC. Add 60 ul ETOH and 2,2 ul 2M ammonium
acetate, pH4.6. Vortex and store at -20 oC >30 minutes or O/N,
(or l5 minutes in dry ice/ethanol bath). Spin l0 minutes and vacuum-dry
the pellet. Resuspend in l0 ul water, add 2 ul annealing buffer
and 2 ul primer. Proceed according the instructions of the sequencing
kit.
Alternative rapid denaturation
& annealing:
5ul DNA, 1ul (EXACTLY) 1N NaOH/0.1mMEDTA and 1 to 5pmol primer,
10ul total volume. Heat at 80-99oC for 5min, (earlier protocols
used 22 or 65 oC). Add (EXACTLY) 4 ul TDMN and leave at R/T for
10 minutes. Use directly in sequencing reaction as annealed template.
TDMN:
0.28M TES (free acid)
0.12M HCl
0.05M DTT
0.08M MgCl2
0.2M NaCl
Note: prepare this and the 1M NaOH solution with maximum precision,
i.e. using graduated flasks.
Ref: Jones, D.S. and Schofield, J.P. NAR vol. 18, No. 24,
7463-64 (with some modifications)
FURTHER PLASMID PURIFICATION
A. On Sucrose Gradients
This method is good for removal of small molecular weight RNA
and high molecular weight E. coli DNA. The plasmid recovered is
usually pure enough for most enzymes.
1. The plasmid from a 1 litre culture is dissolved in 0.5
ml TE.
2. 16 ml 30% sucrose in TE (autoclaved 20 min, 15 psi) is
placed in front chamber of gradient mixer and 18 ml 10% sucrose
in TE (also autoclaved) in back chamber.
3. gradient is then pumped into an SW28 centrifuge tube in
about 10 min, allowing sucrose to run down wall of tube and overlayer.
4. Tube is transferred to rotor bucket and balanced against
a second gradient by removal of some of the upper sucrose layer.
The buckets and rotor are brought to the centrifuge and the plasmid
over layered onto the gradient just before assembling rotor.
5. Gradients are spun at 23,000 rpm for 15 hours at R/T in
Beckman centrifuge, SW28 rotor. (O.K. for 3 to 12 kb plasmids).
6. Gradients are fractionated by displacement with 60% sucrose
in TE (should not be autoclaved) in Isco. 0.5 min fractions are
taken at pumping speed of 3 ml/min and full scale deflection
of 2.0 O.D. (0.5 cm cell is standard but check). Base line must
be set up using H2O or TE and lamp allowed
to warm up for 10 min before this setting made.
7. Expect:
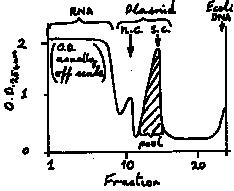
Either
8a. 3 ml of pooled fractions into polyalomel tube for SW28
rotor. Add 1.1 ml of 3 M NaCl and 6.9 ml TE, mix, then fill tube
with cold EtOH and mix again. Leave more than 30 min or O/N at
-20oC.
8b. Dialyse pooled fractions against 2 changes ~100 volumes
each, of TE, over a period of 3 hrs at 4oC.
Finally transfer to polyalomel centrifuge tube (either for SW28
or SW40 rotor) and precipitate with 2 volumes cold EtOH mixing
well. Leave more than 30 min or O/N at -20oC.
9. Recover DNA by centrifuging at 25,000 rpm (SW28 rotor)
or 35,000 rpm (SW40 rotor) and 4oC for
30 min. Pour off supernatant, fill tube with cold 80% EtOH and
spin again for 15 min. Again pour off, supernatant and leave
tubes to drain for a few minutes.
10. Vacuum dry for 5 min and redissolve in TE, ~500 ul or
at 1 ug/ul.
B. On Caesium Chloride-Ethidium
Bromide Gradients
This method gives very pure plasmid DNA as far as enzymes
are concerned. Separation of super-coiled form from nicked circle
form and E. coli DNA is very good, but very little purification
from low molecular weight RNA is obtained as this tends to spread
throughout gradient.
The following is for 10ml or 5ml of final volume.
1. Plasmid from a 1 litre culture is dissolved in ~7ml of
TE, 500ml culture; ~3.5ml.
2. A sterile polypropylene 14 ml tube is placed on a mg accuracy
top-pan balance and exactly 8 g (or 4 gm) CsCl weighed into it.
This is followed, still on the balance, by the 7 ml (3.5 ml)
of plasmid solution and the weight of liquid added made up to
7.5 g (3.75 g) with TE. Wearing disposable gloves 0.5 g (0.25
g) of 10 mg/ml EtBr is added, the tube sealed with parafilm and
the CsCl dissolved thoroughly.
3. Dissolved solution (10ml or 5 ml) is transferred to either
one or two 5ml Quickfit tube(s) (for VTi65 rotor) or to one 13
ml Quickfit tube (for 70.1Ti rotor) and topped up with paraffin
oil using a syringe. Weights of tubes are checked to be within
0.1 g. Seal tubes using the Quickfit sealer.
4. 5ml tubes are centrifuged in VTi65 for 4 hrs at 60krpm
or O/N at 45krpm at 22oC. The 13ml tubes
are centrifuged at 54krpm for 16 hours, or at 35,000 rpm for
more than 48 hours, at 22oC in a 70.1
Ti rotor. Lower speeds give best physical separation as gradient
is shallower. Equilibrium at the lower rpm could be achieved
faster by spinning for 1 or more hours at the higher rpm then
slowing to lower rpm.
5. Tubes are removed from rotor and placed one by one above
the 360 nm UV source.
Expected:
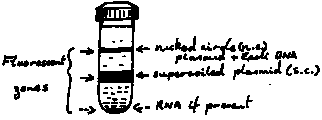
6. Piercing top of Quickfit tube with 18 gauge syringe needle
allows air to enter the tube during extraction. The super-coiled
(sc) zone is extracted from tube by side puncture. A 1 ml syringe
(with 18 gauge needle) is used to puncture the tube just below
the sc zone and the zone drawn into the syringe. NOT more than
1 ml should be taken.
7. Transfer this 1 ml of supercoiled plasmid to a 4 ml polypropylene
tube.
8. Add 1 ml of isoamyl alcohol and shake well. Remove upper
phase and discard. Repeat this step once, twice or more times
until most of the EtBr is removed from plasmid.
9. Transfer plasmid to polyalomel SW28 centrifuge tube or
30 ml polypropylene tube. Add 10 ml TE, mix, then fill tube with
cold EtOH, mix again and leave at -20oC
for more than 30 min. but not O/N.
10. Recover plasmid by centrifuging at 25,000 rpm for 30 min
and 4oC in Beckman, SW28 rotor or at 10,000
rpm for 30 min and 4oC in Sorvall RC-5B,
HB4 rotor. Pour off supernatant, fill tube with cold 80% EtOH
and spin again for 15 min. if necessary. Again pour off supernatant
and leave tubes to drain for a few minutes.
11. Vacuum dry for 5 min, (NOT for cosmids, which should be
just left to drain) and redissolve in TE - about 500 ul for plasmids,
100 ul for cosmids (~1 ug/ul).

